Next generation sequencing (NGS) has shown great potential in the detection and discovery of novel animal/plant pathogens. In this regard, it is common to distinguish between the spread of known infections to new areas and/or the emergence of completely novel, ‘unknown’ pathogens. Contrary to earlier techniques, NGS is unbiased and reports all nucleotide sequences present in the original sample. However, as with earlier techniques, the lower limit for detection is still ultimately determined by the abundance of pathogens in relation to host background material. By enabling deeper sequencing to be performed more quickly and cheaply, the continuing development of NGS techniques is also continuing to improve the likelihood of detecting low copy-number pathogens.
Case study
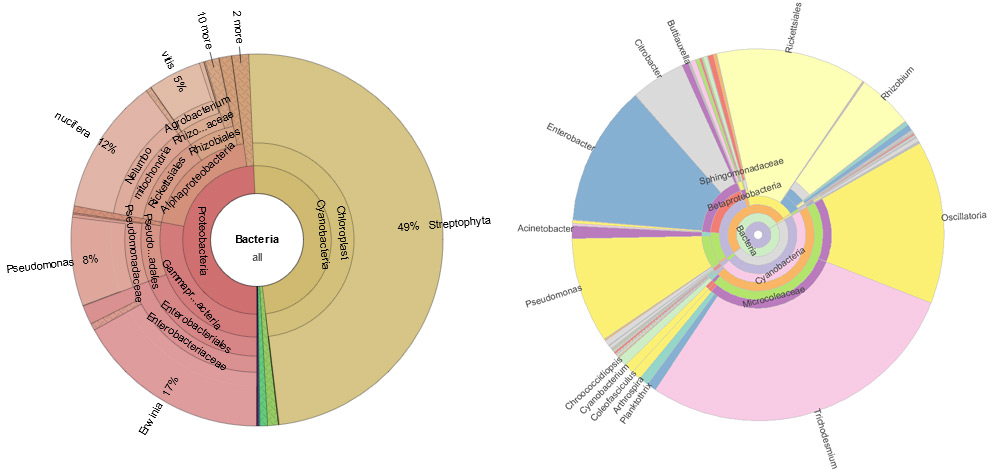
Effects of crown gall disease on natural microbiota of Vitis vinifera: reanalyzing EBI Project ERP013477, sample ERR1171009
The crown gall disease in grapevine is mediated by Agrobacterium vitis and results in tumorous growth. The tumor offers an ecological niche for the pathogen and eventually for other bacteria as well. Using next generation sequencing of the V4 region of 16S rDNA amplicons we recorded the microbiota from galled and non-galled grapevine plants as well as of the soil at three seasons of the year. The root, trunk, one-year-old cane and the soil hosted distinct microbiota, which differed according to spring, summer, and autumn. However, the crown gall disease influenced the structure of the microbiota only in grapevine trunks. There the bacterial richness was higher and comprised for example exclusively the plant pathogens Xanthomonas sp., Sodalis sp., and an unknown Gammaproteobacterium. The seasons additionally affected the microbiota in the trunks with and without a crown gall differently. A. vitis, Pseudomonas sp. and Enterobacteriaceae sp. were the most abundant sequences in crown galls at each season. In contrast, in healthy trunks (=non-galled) the microbial composition changed from season to season. Altogether, twenty-two operational taxonomic units, representing bacterial species, were significantly enriched in crown galls and only three in healthy trunks. The results support the idea that other bacteria than the pathogen A. vitis can adapt to the crown gall environment.
Figure 1. Seqomics' reanalyzation shows updated view of bacterial taxas