~99% of identified prokaryotes from the marine environment, cannot be cultured in the laboratory. This lack of capability restricts our knowledge of microbial genetics and community ecology. Metagenomics has already provided a wealth of information about the uncultured microbial world. It has also facilitated the discovery of novel biocatalysts by allowing researchers to probe directly into a huge diversity of enzymes within natural microbial communities. Recent advances have led to a great interest in recruiting microbial enzymes for the development of environmentally-friendly industry. Metagenomics approach expected to provide not only scientific insights but also economic benefits, especially in industry.
Case study
A metagenomics transect into the deepest point of the Baltic Sea reveals clear stratification of microbial functional capacities: reanalyzing EBI Project ERP002477, sample ERR268109
The Baltic Sea is characterized by hyposaline surface waters, hypoxic and anoxic deep waters and sediments. These conditions, which in turn lead to a steep oxygen gradient, are particularly evident at Landsort Deep in the Baltic Proper. Reconstruction of the nitrogen cycle here shows potential for syntrophy between archaeal ammonium oxidizers and bacterial denitrification at anoxic depths, while anaerobic ammonium oxidation genes are absent, despite substantial ammonium levels below the chemocline. There's an enrichment in genetic prerequisites for a copiotrophic lifestyle and resistance mechanisms reflecting adaptation to prevalent eutrophic conditions and the accumulation of environmental pollutants resulting from ongoing anthropogenic pressures in the Baltic Sea.
Figure 1. Krona graph of highly diversed and unclassified, novel taxons
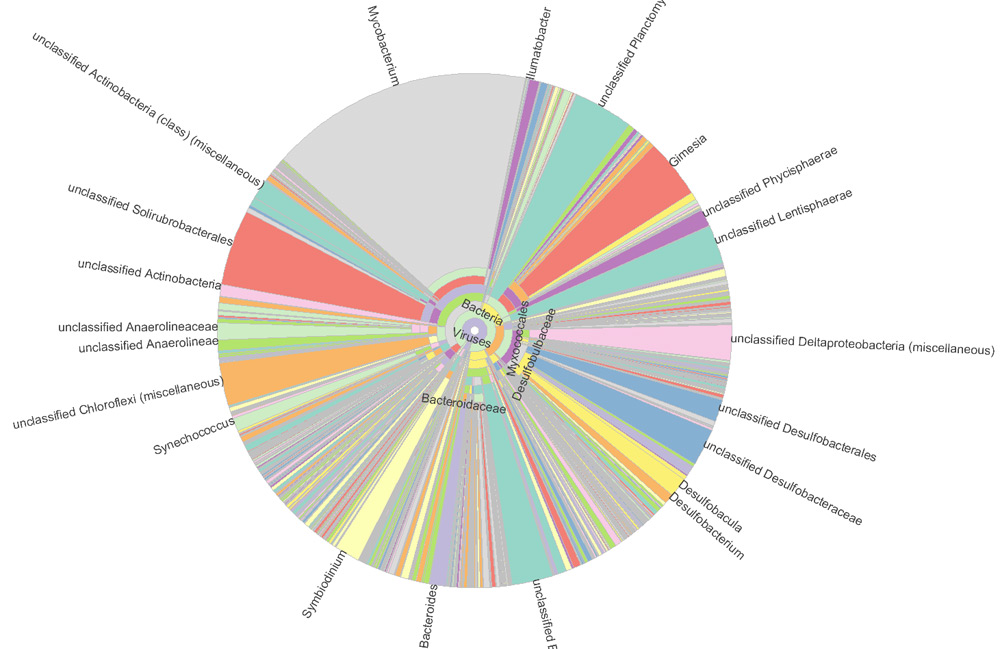
Figure 2. Krona graph of the viral clade with the assigned number of reads
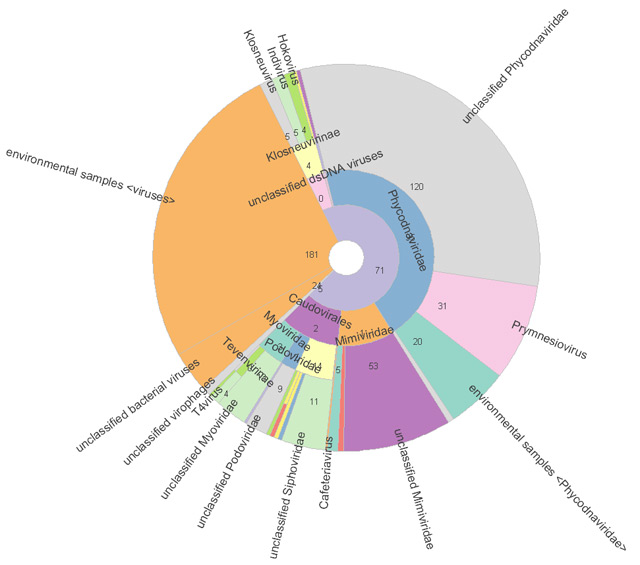
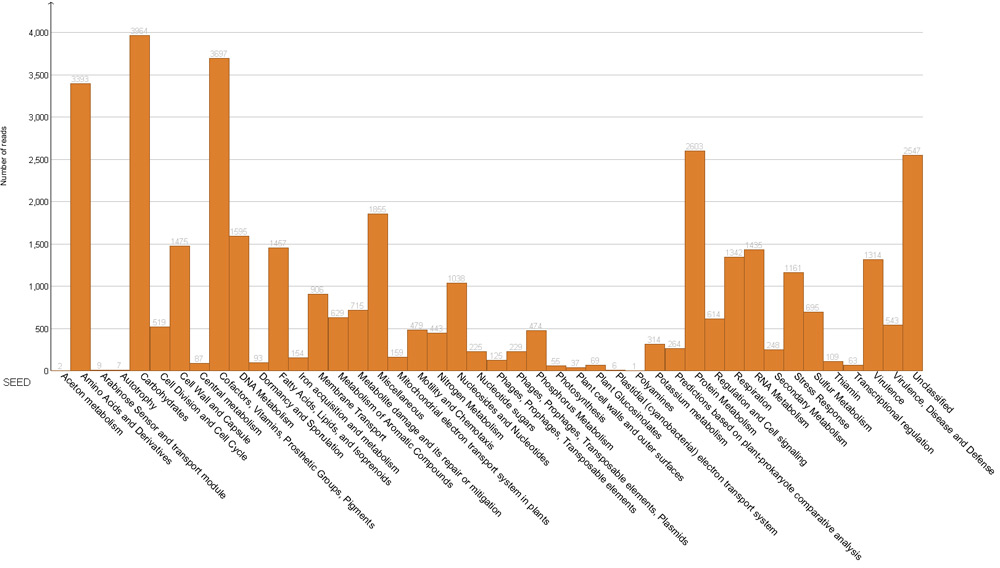
Figure 3. Functional analysis based on main SEED categories