Differentiating between infectious and non-infectious illness, and identifying the underlying etiology of infection, can be quite challenging. For example, more than half of cases of encephalitis remain undiagnosed, despite extensive testing using state-of-the-art clinical laboratory methods. Metagenomic sequencing shows promise as a sensitive (as few as one read depth) and rapid method to diagnose infection by comparing DNA found in a patient's sample to the latest non-redundant database of NCBI.
Case study 1.
Links
SRR5945841.daa (results in MEGAN6 compatible file, please extract with WinRAR)
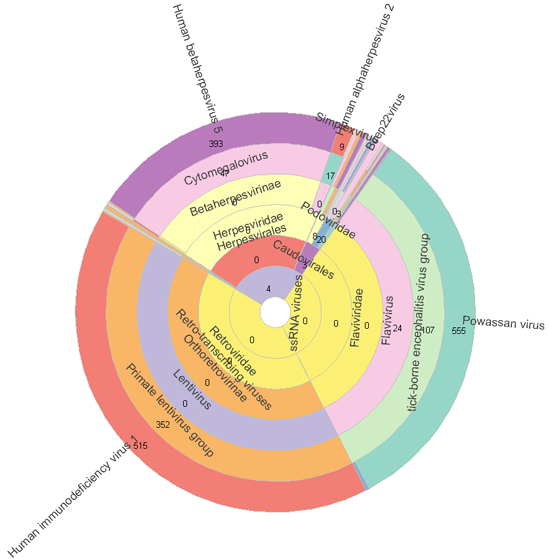
Case study 2.
Southampton Asthma metagenomics: reanalyzing EBI project ERP006003, sample ERR525606
55 subjects underwent detailed clinical and immunological phenotyping, sputum induction and bronchoscopy during periods of clinical stability. In addition to 15 healthy controls, 9 mild, 16 moderate and 15 severe ashthmatics (without bronchiectasis) were included. Protected bronchoalveolar lavage (BAL) and induced symptoms were analysed by WGS for RNA and DNA from bacterial, viral and fungal genomes. 88 samples were sequenced using 454 FLX Titanium. Single end library. Data were provided as fasta files following adapter trimming.
Figure 2. Analysis reveals massive amounts of Streptococcus and Prevotella related sequences